Complete Solution
🚀 From Computer Vision to Production
This project demonstrates the complete workflow of an ML system: from data labeling and model training to cloud deployment and monitoring. Not a "proof of concept" but a working system that NPEC researchers could use.
System Features
🔬 Accurate Detection
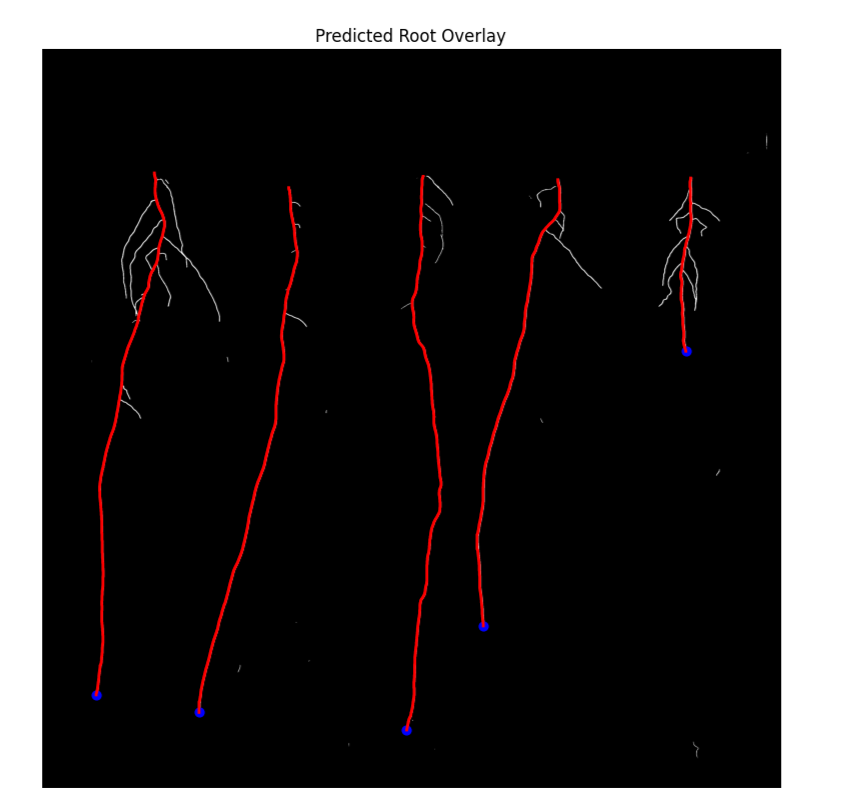
U-Net architecture for root detection with 11% SMAPE score
📊 Per-Root Tracking
Track up to 5 plants simultaneously with measurements and growth rate per individual root
🤖 Robot Integration
Exact root-tip coordinates for automated feeding system
📈 Growth Analysis
Visualizations of root growth over time with trend analysis per plant
☁️ Cloud Deployment
Azure Container Apps with auto-scaling and 24/7 availability
🔄 Auto-Training
Automatic model updates with new data via feedback loop